Welcome to m6AConquer !
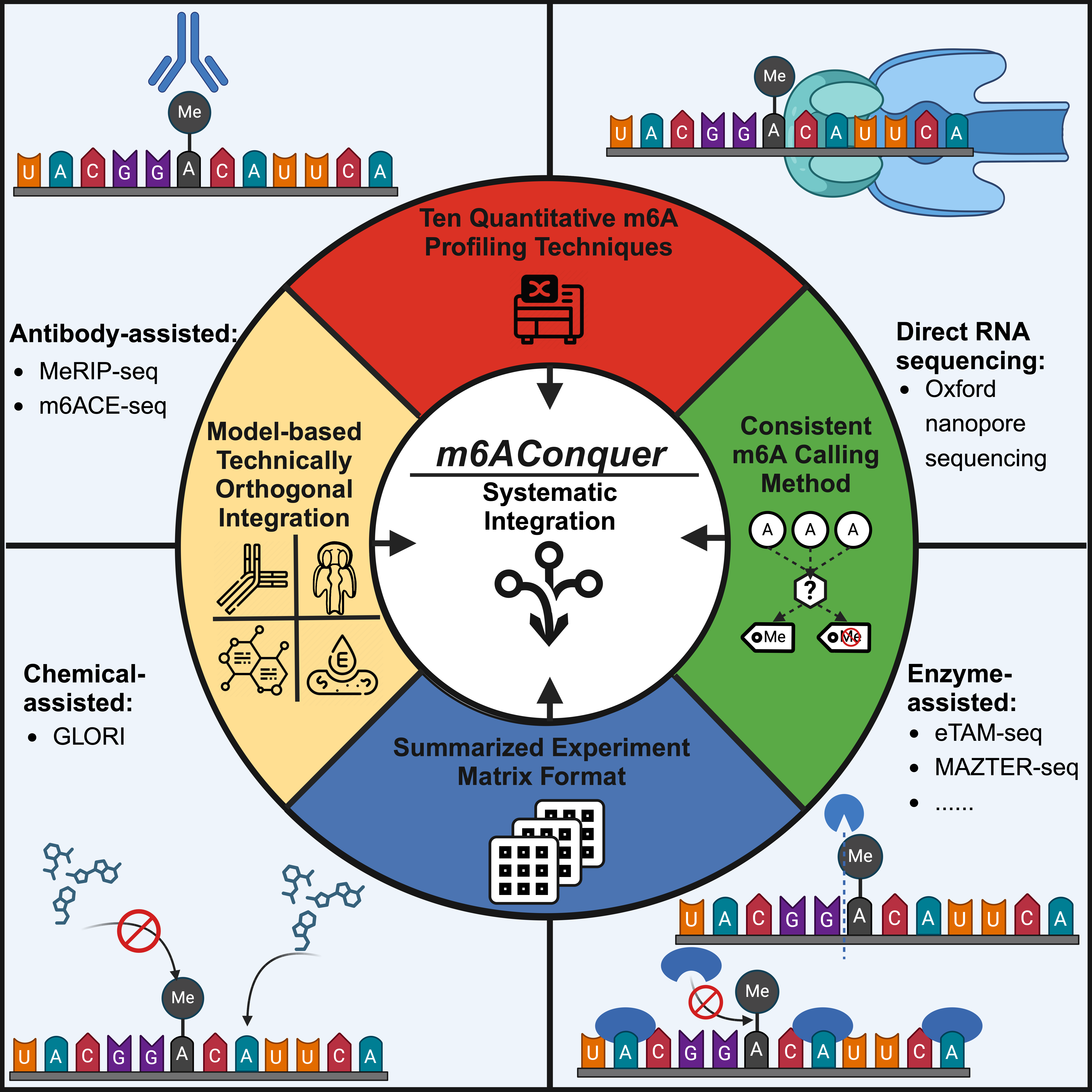
N6-methyladenosine (m6A) is the most prevalent RNA modification in mammalian cells and the most extensively studied epitranscriptomic mark. The m6AConquer project (Consistent Quantification of External m6A RNA Modification Data) summarizes quantitative m6A data from 10 detection techniques using 2,788,705 (human) and 2,761,121 (mouse) consistent base-resolution features.
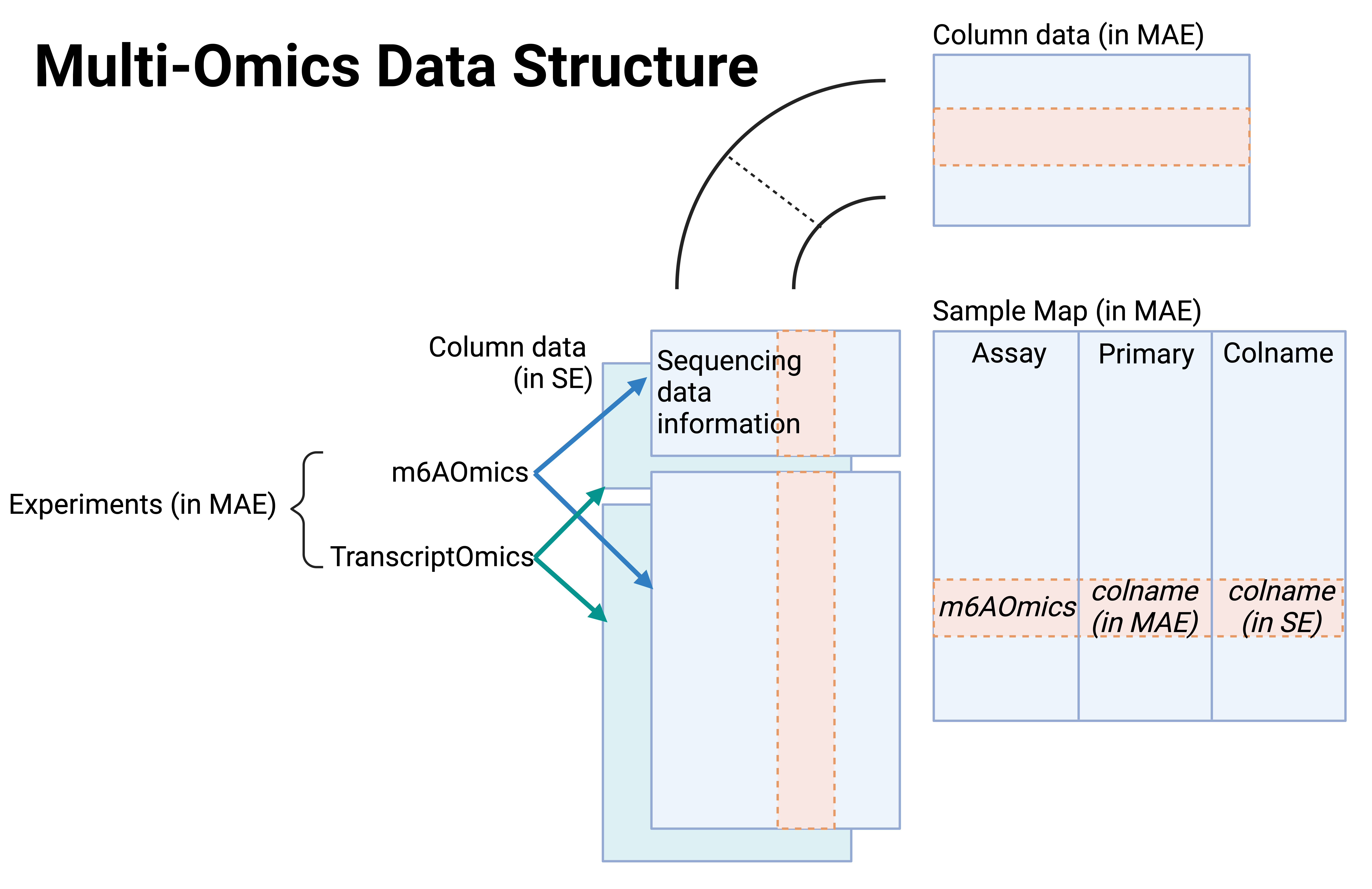
Available m6A detection techniques fall into 4 categories: antibody-assisted, chemical-assisted, enzyme-assisted, and direct-RNA sequencing. m6AConquer leverages this categorization to integrate and identify 139,700 orthogonally validated m6A sites reproducible across independent techniques in human and mouse. Additionally, m6AConquer consistently quantifies m6A profiling data and unifies site calling procedures, providing a standardised data-sharing framework. 4 types of multi-omics data, including m6A methylomes, expression levels, alternative splicing events, and variant calling results, are embedded into this framework. Using m6A methylome and genetic variant data, m6AConquer provides 6,034 m6A QTL locations. The current version of m6AConquer archived 1,928 FASTQ and FAST5 data, 1,394 sequencing samples, 72 cell lines or tissues, and 13 integrated orthogonal technique pairs.
Query 
Query m6A sites by gene names
Query m6A modification levels by multiple genes
Integration 
Visualization of reproducibility-based integration and browse orthogonally validated m6A sites
In-depth analysis of regulatory contexts of orthogonally validated m6A sites via UMAP
m6A QTL 
Browse m6A quantitative trait locus and disease annotation
Download 
Download quantitative data sharing frameworks
Download sample specific data quality control report
Download m6A sites validated by orthogonal techniques
Download orthogonally validated differential m6A sites
Download m6A quantitative trait locus
IGV Browser 
Browse m6A sites validated by orthogonal detection techniques
Help 
User guides on how to use m6AConquer website
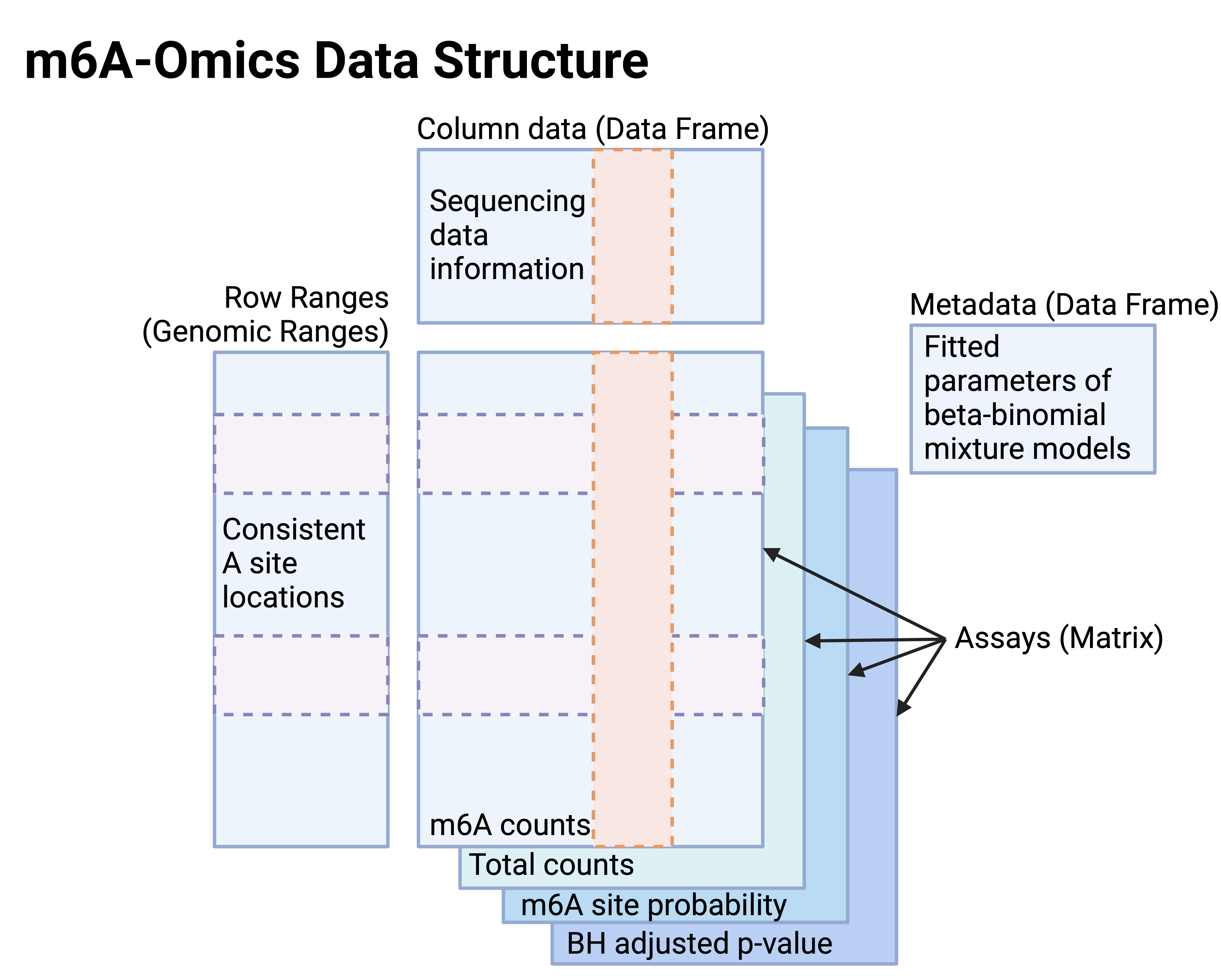
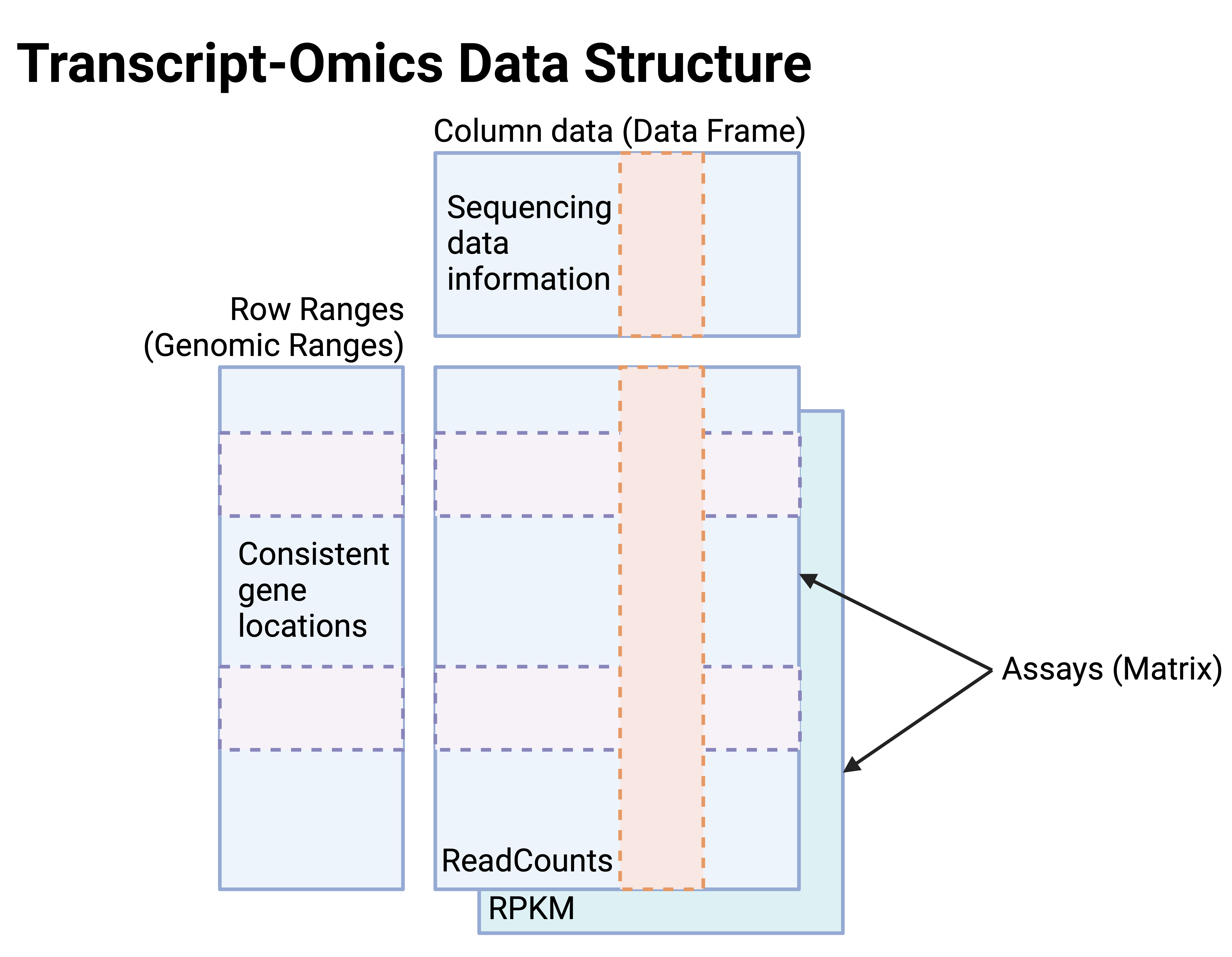
User guides on how to use single-omics, multi-omics, and orthogonal validated m6A site data frameworks
Data resources used in m6AConquer project and database construction pipelines
m6AConquer Release Note:
- Update m6AOmics SE and MAE objects
- Add high and low stringency options for m6A QTL module
- Add downloadable benchmark dataset & supplementary rowrange data
- Add downloadable 0-based BED files
- Add interactive UMAP analysis
- Add downloadable OmicsFeature for human and mouse
- Add alternative splicing and SNP data matrices
- Add m6A QTL module
- m6AConquer is online.