The cross-technique integration is performed using IDR (Irreproducible Discovery Rate) analysis. IDR is well-established in DNA epigenetics and used by the ENCODE database to integrate CHIP-Seq experiments. The IDR statistical framework effectively uses rank information to quantify consistency and eliminate spurious overlap between replicated omics experiments. Because the measure of consistency is not confounded by platform-dependent thresholds, inter-platform consistency can be assessed easily.
In m6AConquer, the reproducible sites identified by IDR < 0.05 are both ranked highly based on their m6A levels (m6A/Total Coverage) and show a positive correlation in m6A levels across paired m6A detection techniques. In this page, a table will be returned after the query to display reproducible sites filtered with an IDR < 0.05.
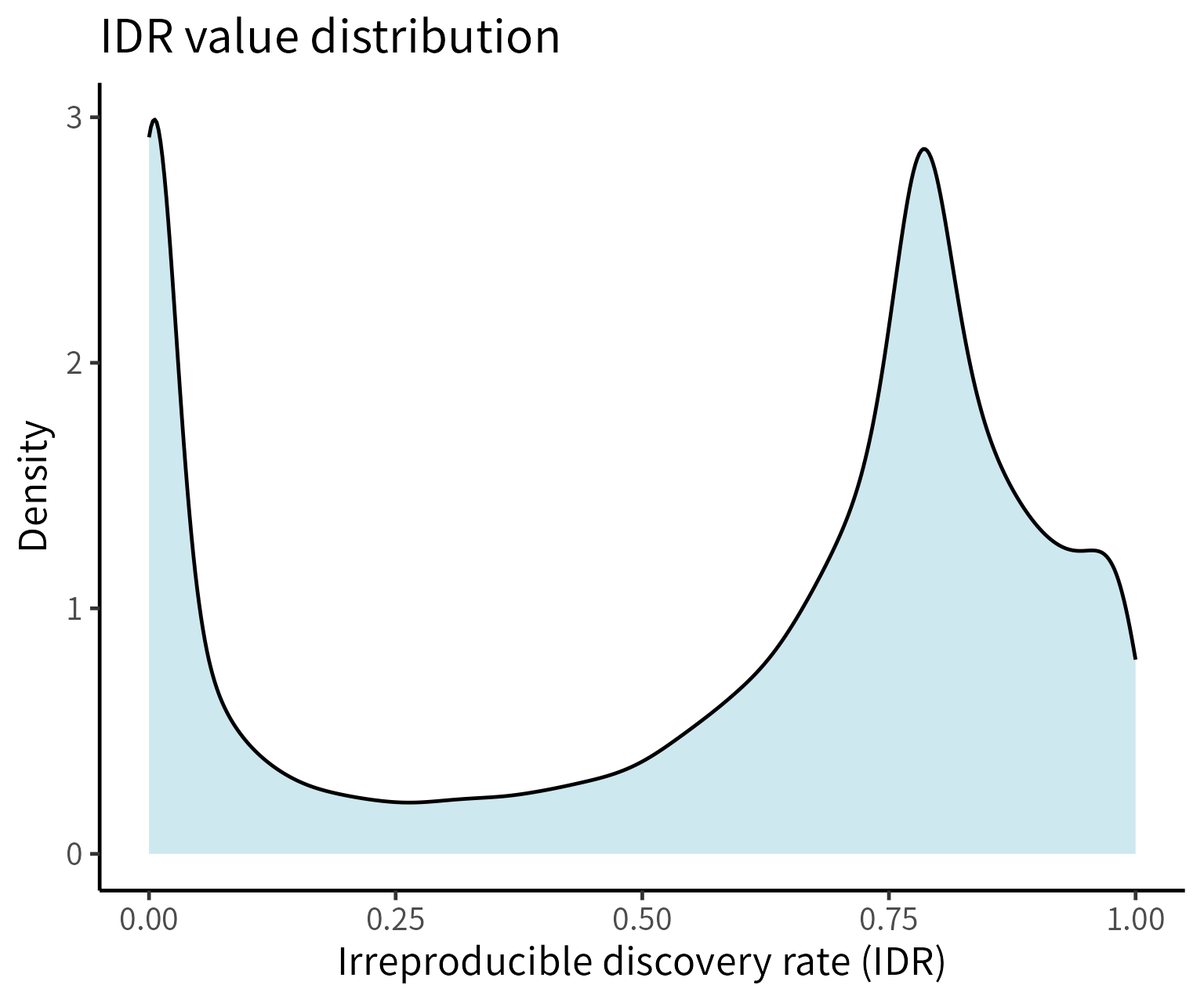
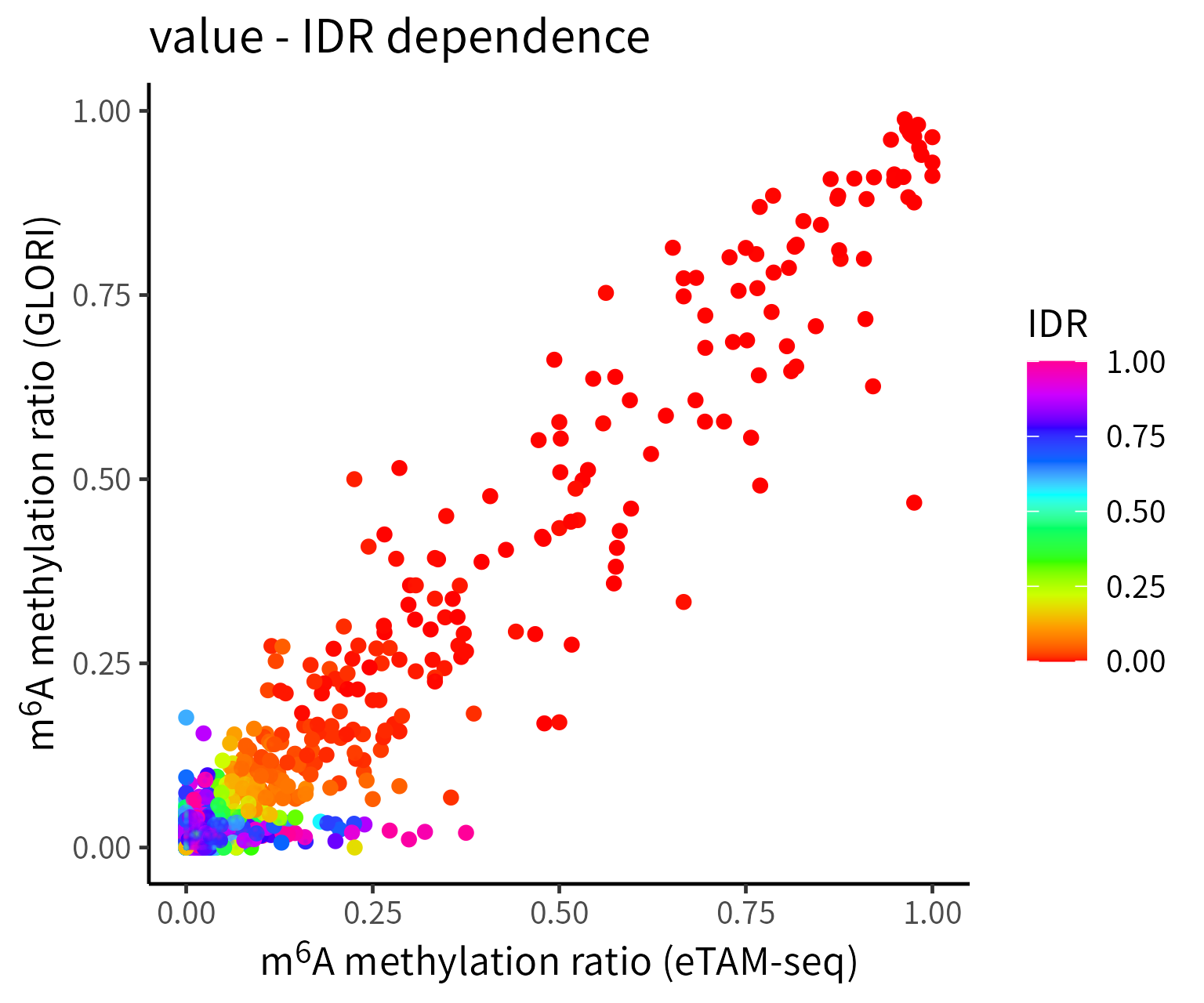
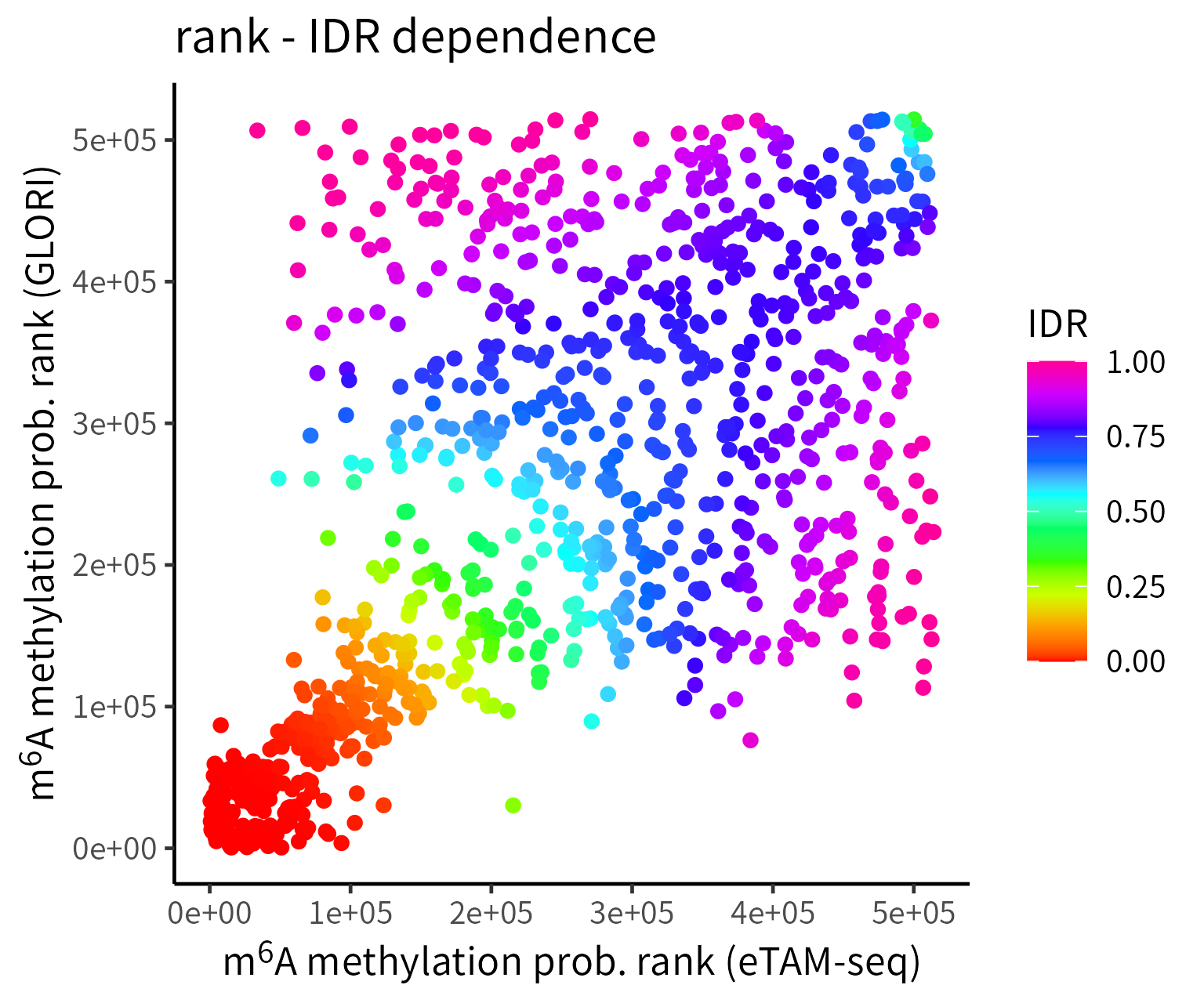
Description of Figures in IDR Analysis:
- Scatter plots of rank: The axes represent the ranks of m6A level in each technique, while the color indicates IDR values computed at the candidate m6A sites.
- Scatter plots of ratio: The axes represent the m6A level in each technique, while the color indicates IDR values computed at the candidate m6A sites.
- Distribution of IDR: The distribution of IDR values for a pair of m6A detection techniques. The density closer to the left indicates reproducible sites, while the density farther from the left indicates background sites.
Reproducible m6A Sites
| Site ID | Chromosome | Position | Strand | IDR | m6A Ratio | m6A Posterior Prob | BH Adjusted P-Value | Technique | JBrowse |
|---|