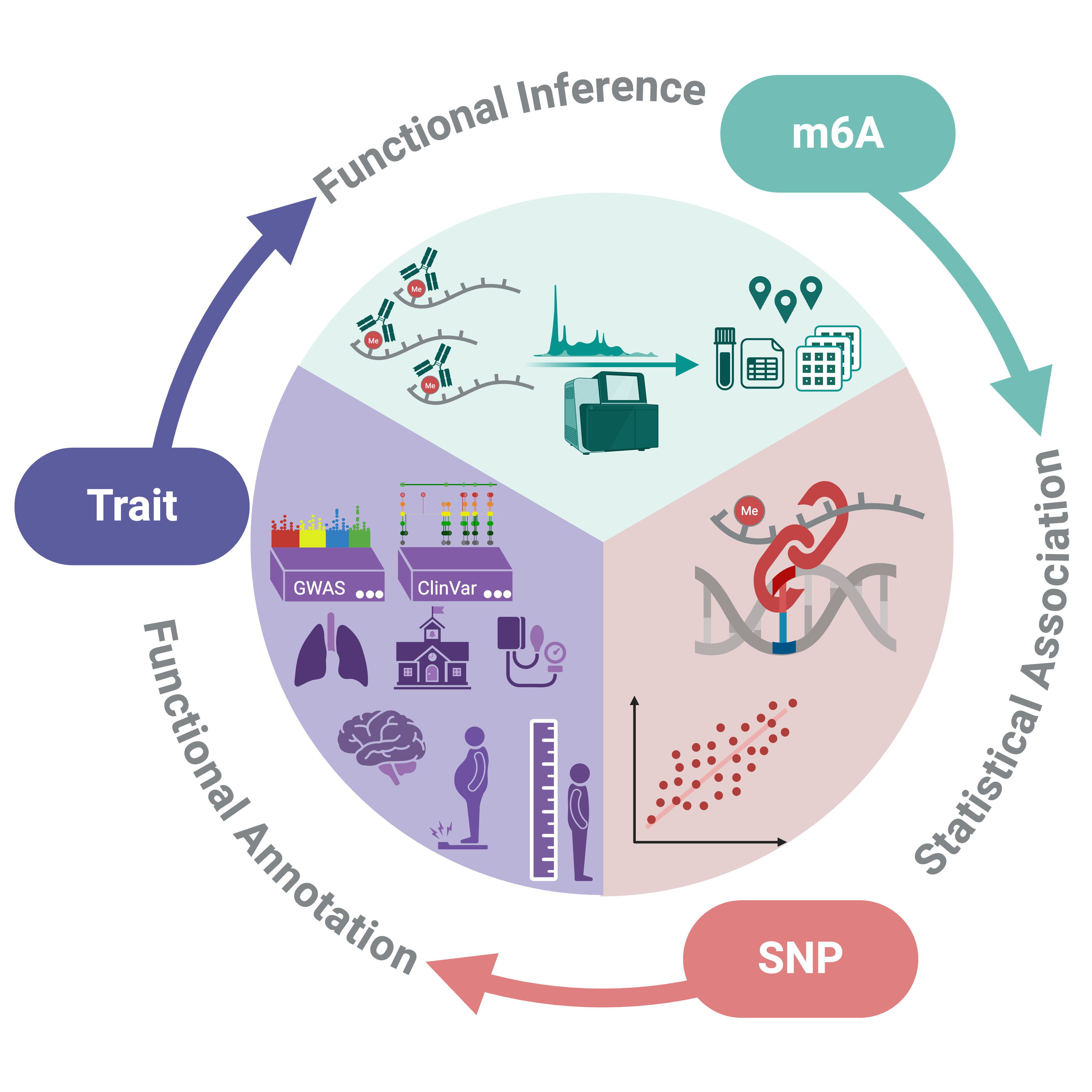
m6A QTLs (m6A Quantitative Trait Loci) are genetic variants (e.g., SNPs) associated with variation in N6-methyladenosine (m6A) methylation levels on RNA. These associations may influence RNA stability, translation, and disease risk, but require experimental validation to establish causality. In m6AConquer, we performed m6A QTL analysis on orthogonally validated human sites, using consistently quantified MeRIP-seq data. The identified m6A QTLs were functionally annotated by GWAS and ClinVar databases.
In this page, you can query the m6A QTLs in m6AConquer by filtering nominal P-Value, empirical P-Value, and disease annotation from GWAS or ClinVar.
- Nominal P-Value: The raw, uncorrected p-value from a single statistical test of association between a SNP and the methylation level of a m6A site.
- Empirical P-Value: The beta distribution-adjusted p-value that accounts for the multiple SNPs tested for each m6A site, thereby improving result reliability.
To assess the quality of our m6A QTL dataset:
- We analyzed variants located within the DRACH motif of their associated m6A site (empirical p-value < 0.1).
- Variants that disrupt the DRACH motifs have exclusively negative effect sizes (slopes), which are significantly more negative than those of non-disrupting variants (t-test p = 0.00775).
- This concordance with biological mechanisms validates the high accuracy of our data.

m6A QTL Sites
| Site ID | Chromosome | Position | Strand | Gene Symbol | Dist2Junc | Variant ID | Nominal P | Slope | Slope SE | Empirical P | GWAS | ClinVar |
|---|