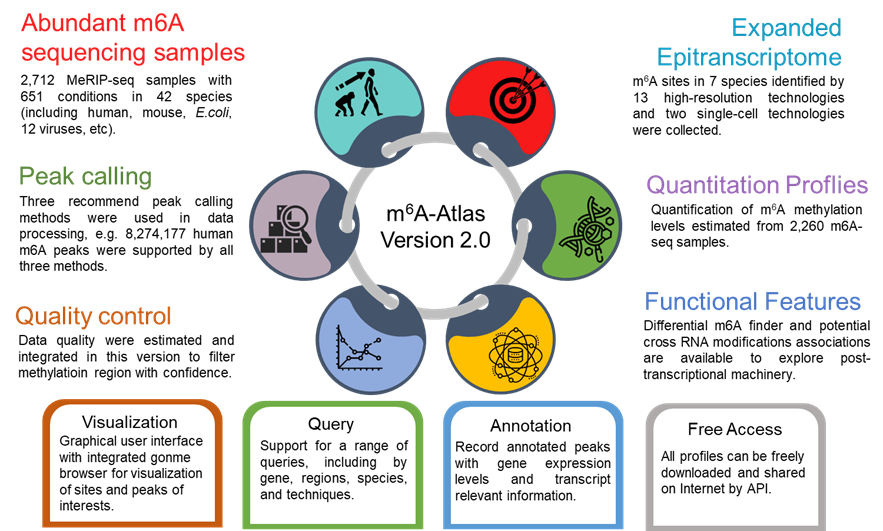
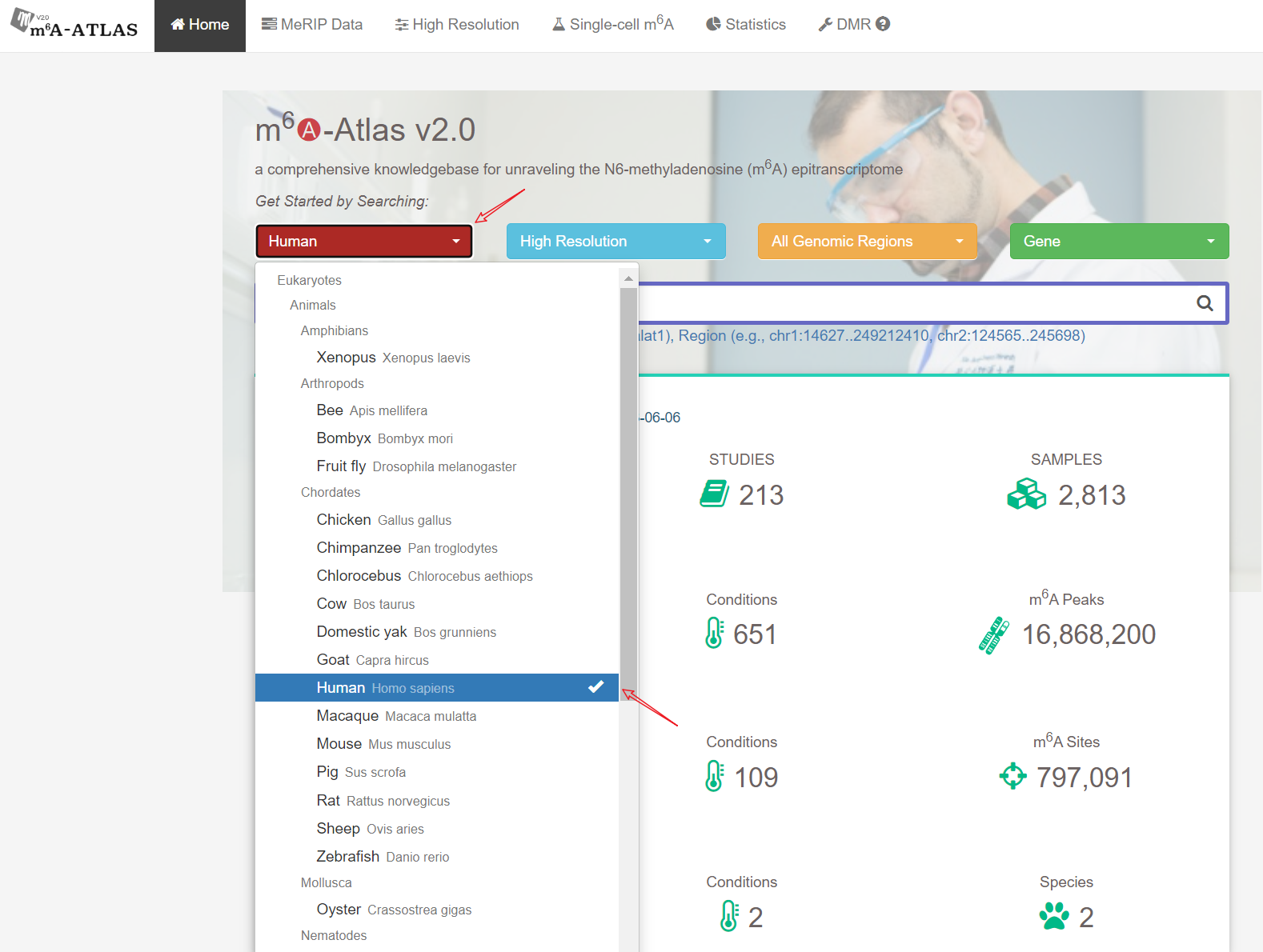
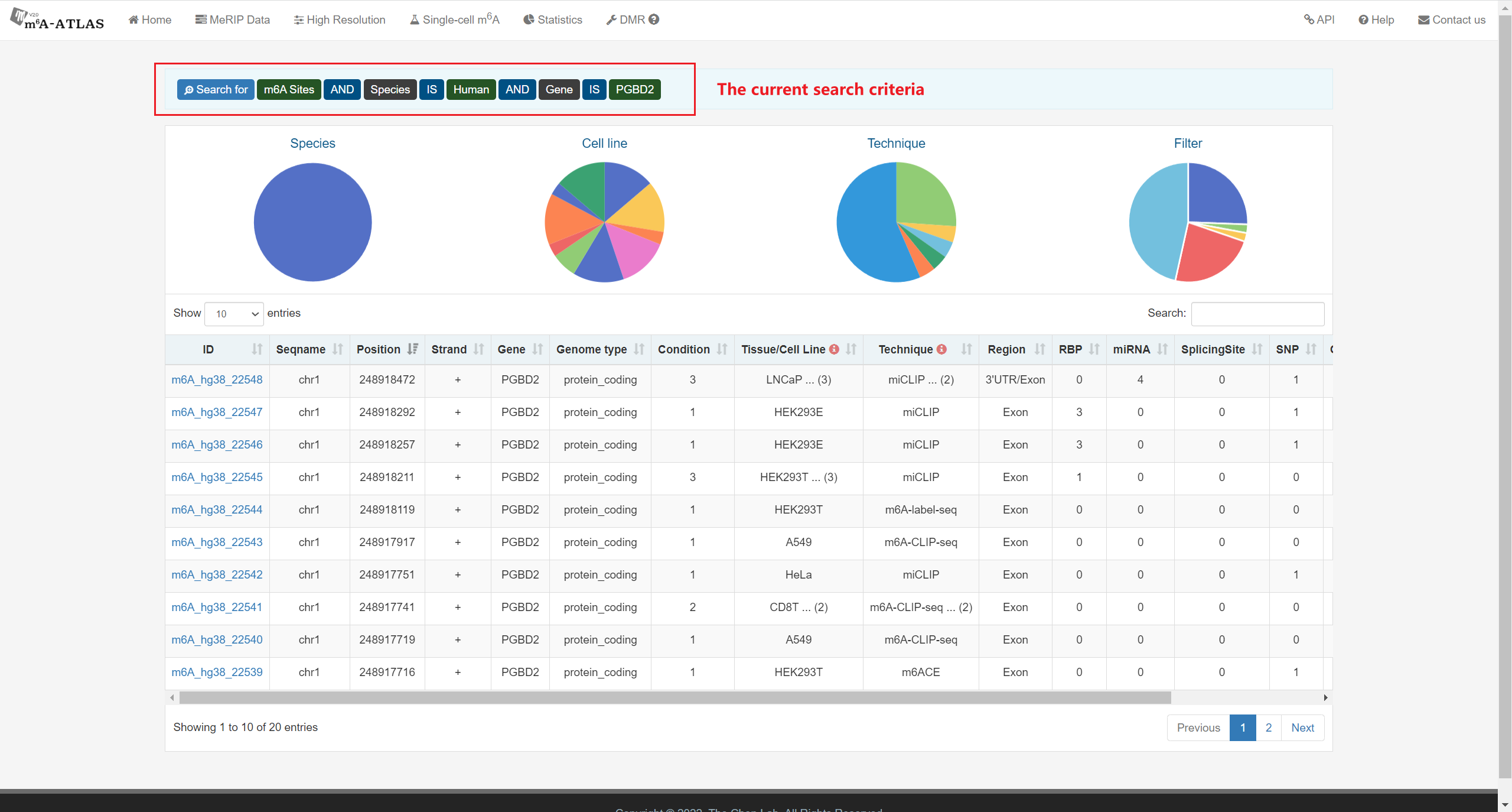
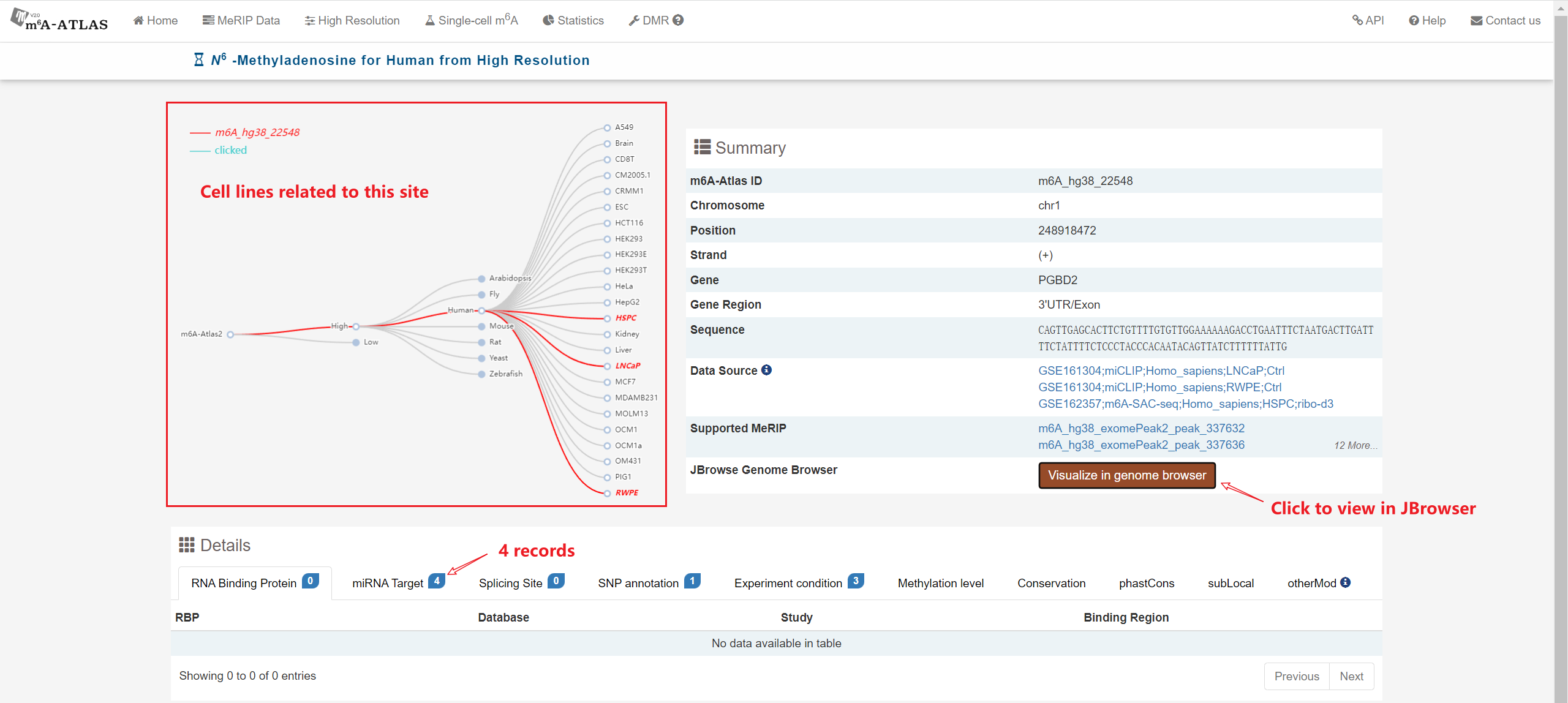
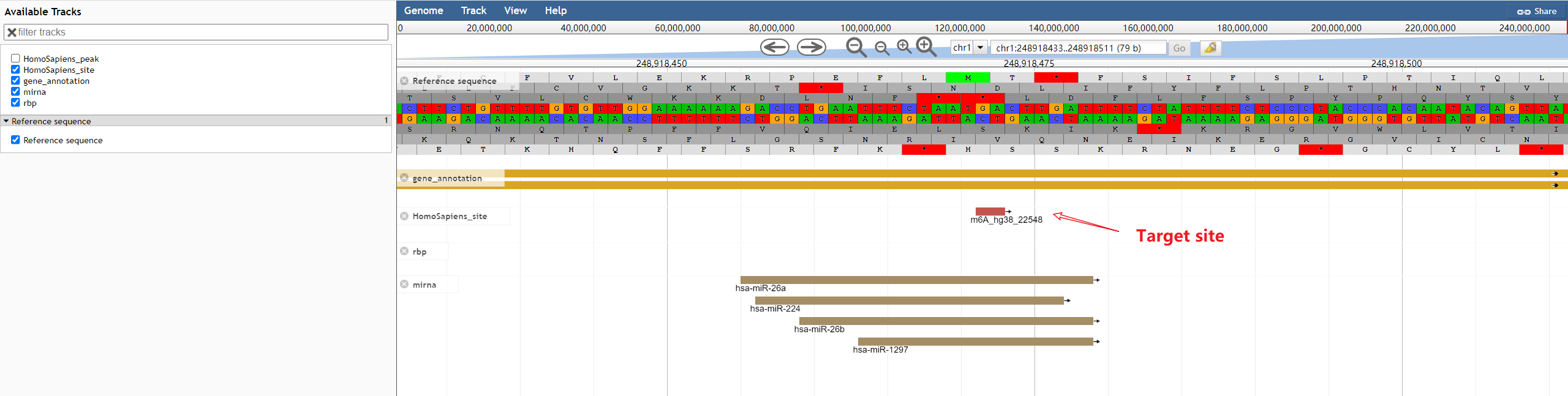
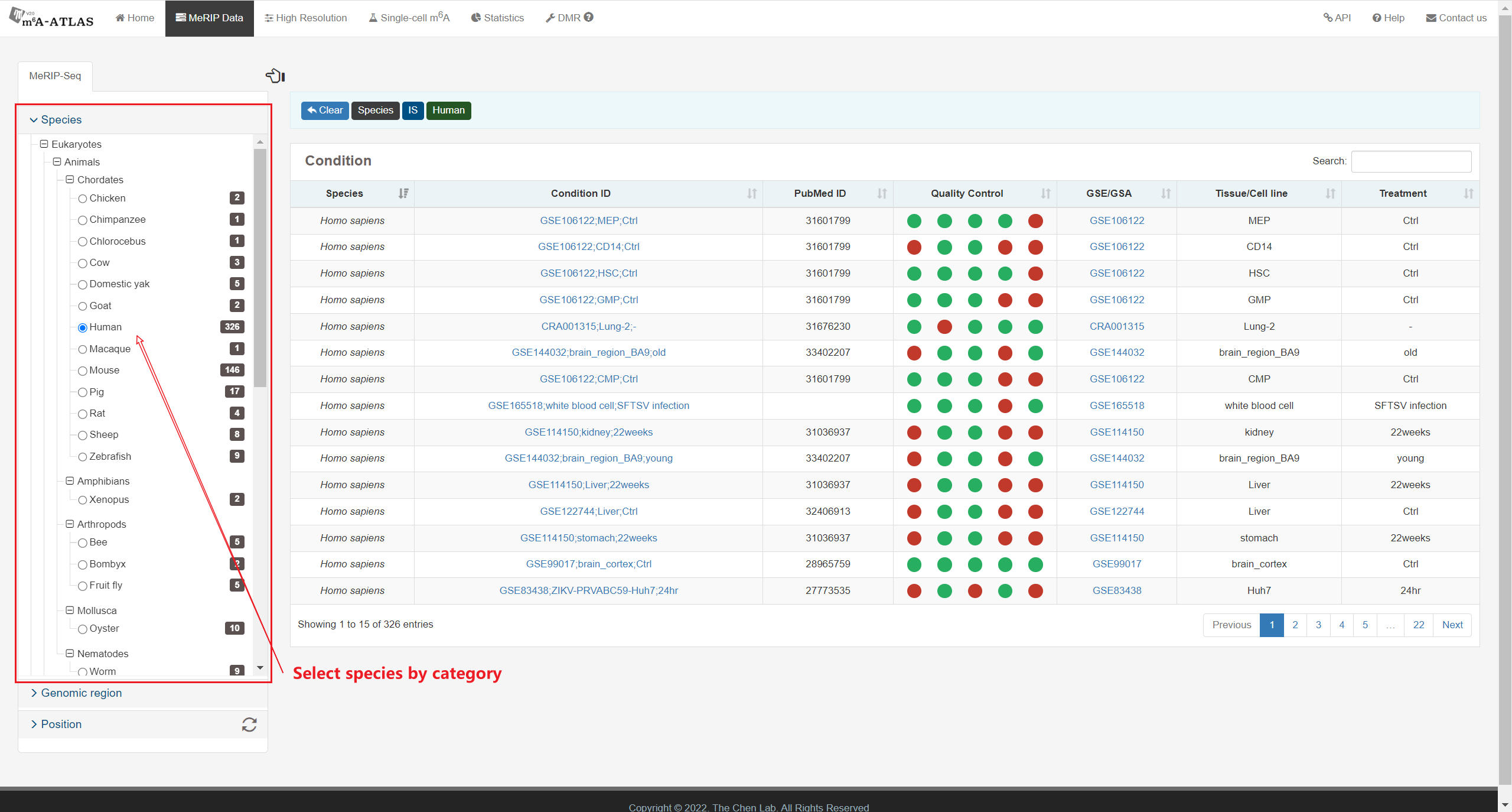
m6A-Atlas v2.0 was expanded to include 797,091 reliable m6A sites among 7 species, with 13 high-resolution technologies and 109 conditions. Additionally, sites derived from single-cell technique were presented for the first time. It estimated quantitative epitranscriptome profiles under 241 conditions for human and 129 conditions for mouse. A user-friendly graphical interface was constructed to support the query, exploration and sharing of the m6A epitranscriptomes annotated with putative post-transcriptional machinery (SNP association, conservation, RBP-binding, microRNA interaction, splicing sites, subcellular location and circRNA generation).
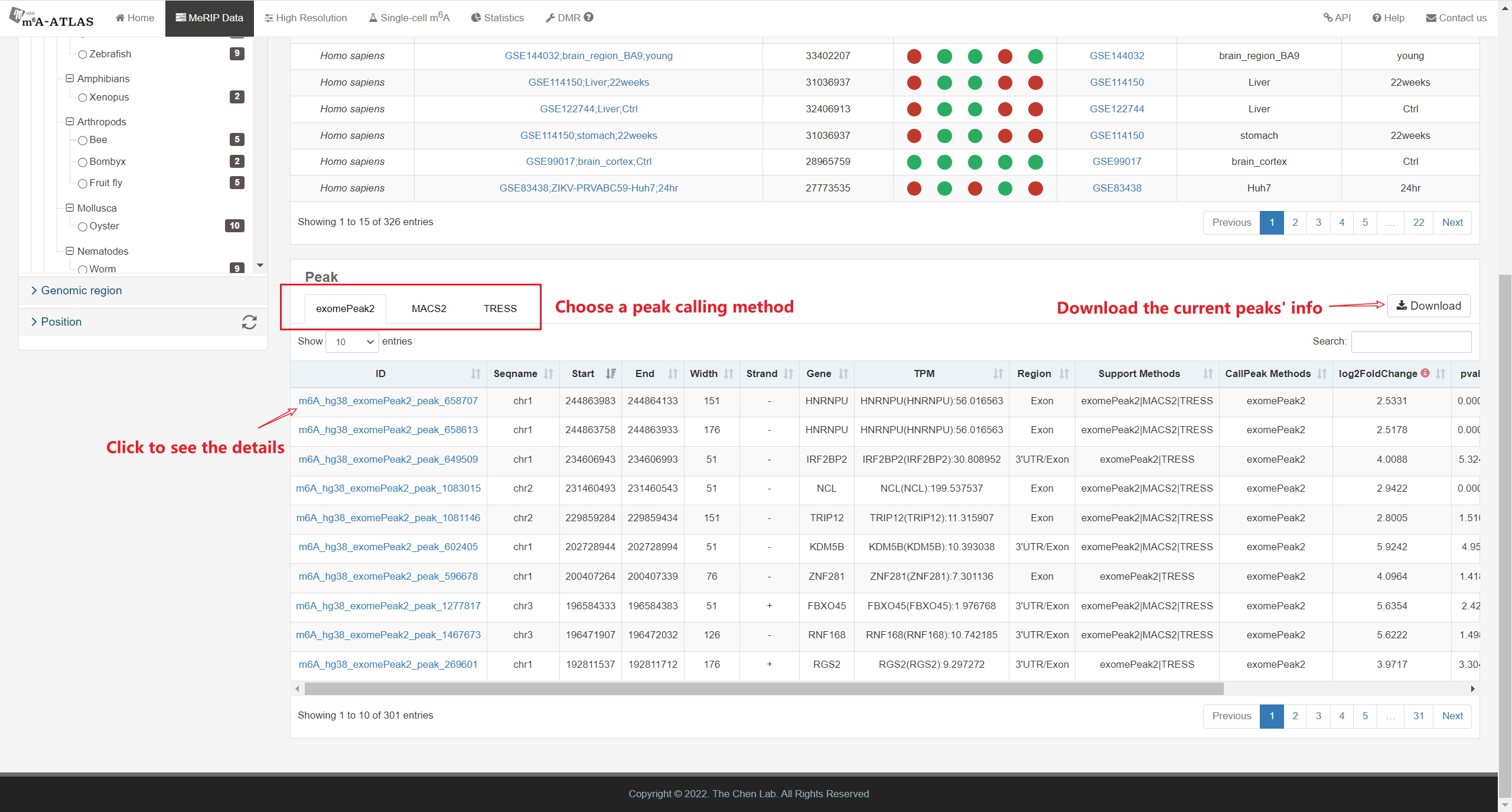
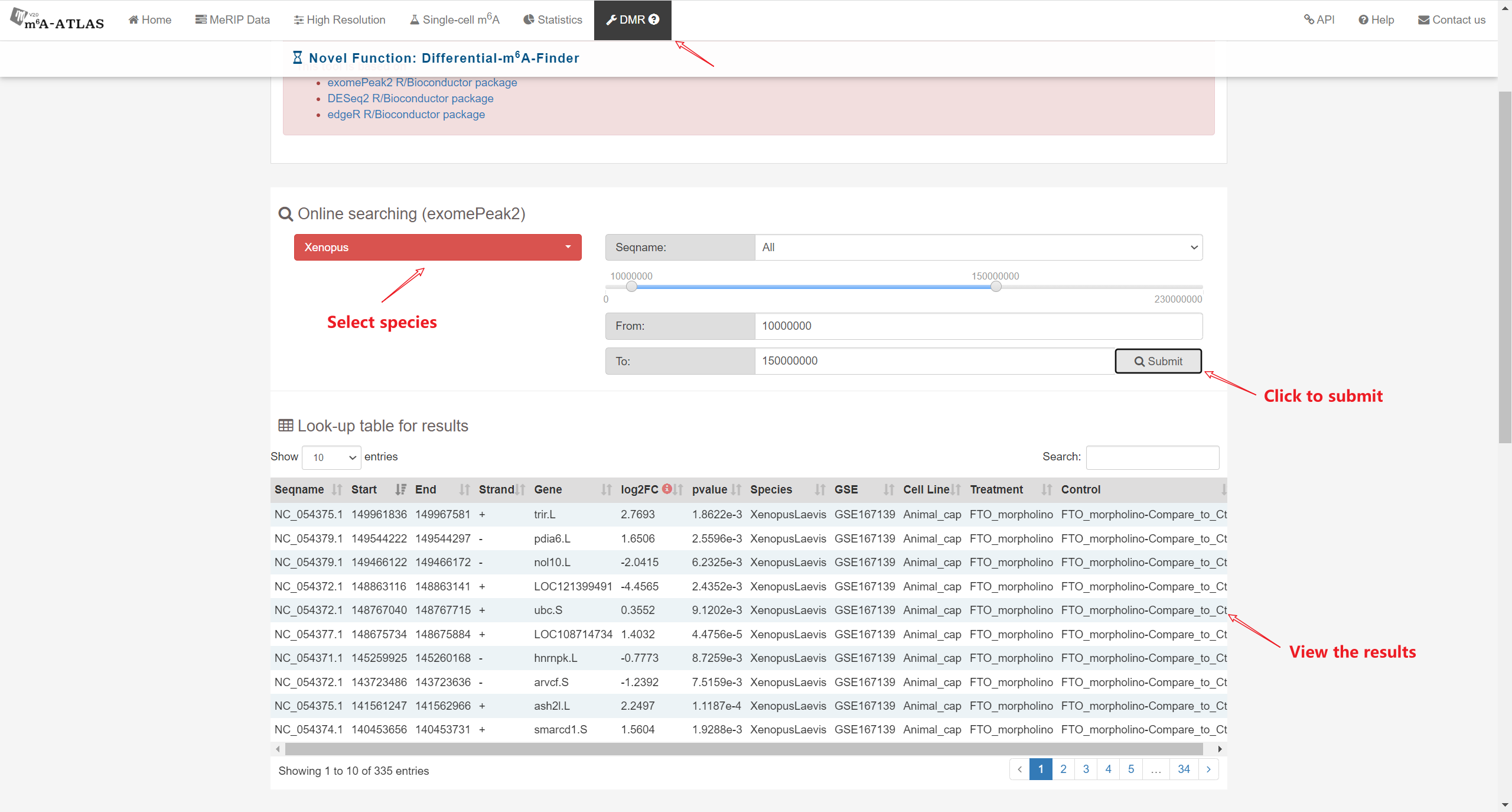
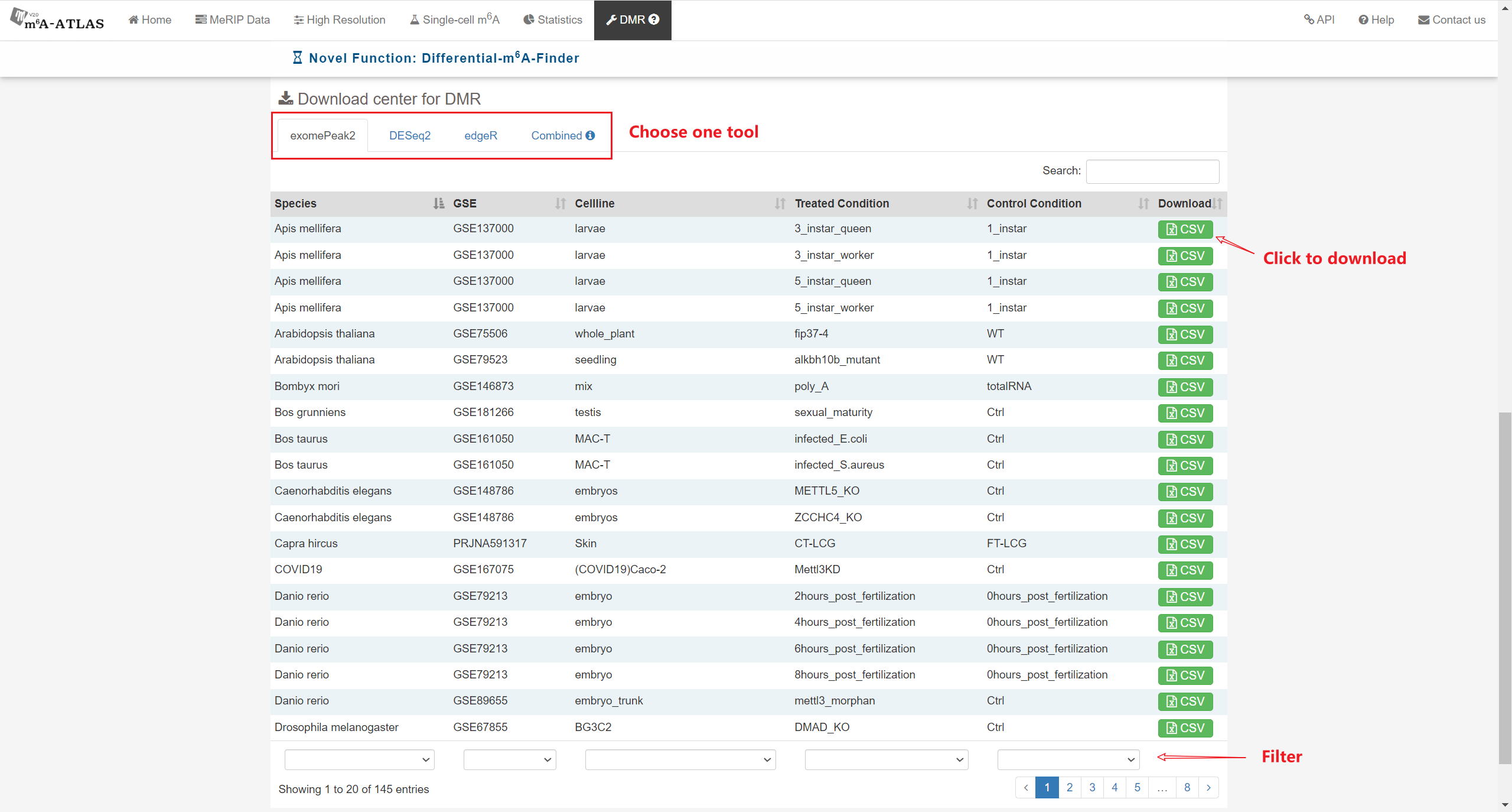
Besides high-resolution data, 16,868,200 m6A enriched regions were recorded in the updated version from 2,712 MeRIP-seq samples. Compared with other RNA modification databases, m6A-Atlas v2.0 covers the most species (30 species and 12 viruses in Atlas v2.0 VS 12 species in RMBase 2.0). We also provide a novel function that enables querying of differentially methylated m6A peaks between different conditions.