Introduction
Since the first post-transcriptional modification was identified nearly 60 years ago, more than 170 chemical modifications have been documented on RNA. 5-methylcytosine (m5C) is one of the most prevalentmodifications, which can be found on both coding and non-coding RNAs and distributes in different species. High throughput sequencing technologies, and couples to specific antibodies or chemical treatment, enable the comprehensive study of m5C epitranscriptome. To integrate the results of various experiments, we built a database, called m5C-Atlas, to summarize candidate m5C sitesdiscovered by five technologies (i.e., BS-seq, miCLIP, RBS-seq, m5C meRIP and Aza-IP) in twelve species (i.e., Homo sapiens, Arabidopsis, Brassica rapa, Caenorhabditis elegans, Danio rerio, Drosophila melanogaster, Ginkgo biloba, Murine leukemia virus, Mus musculus, Saccharomyces cerevisiae, Triticum turgidum subsp and Nannochloropsis oculata). Data from BS-seq technologies was processed to address high confident modified sites. Site information, including gene region, whether it is spliced, whether it is targeted by miRNA, and whether it is a SNP site, were annotated.===============================================
User guide
1.Query by genomic coordinates or gene name1)m5C-Atlas provides users a function to query possible m5C sites withingenes or specific regions in the genome.
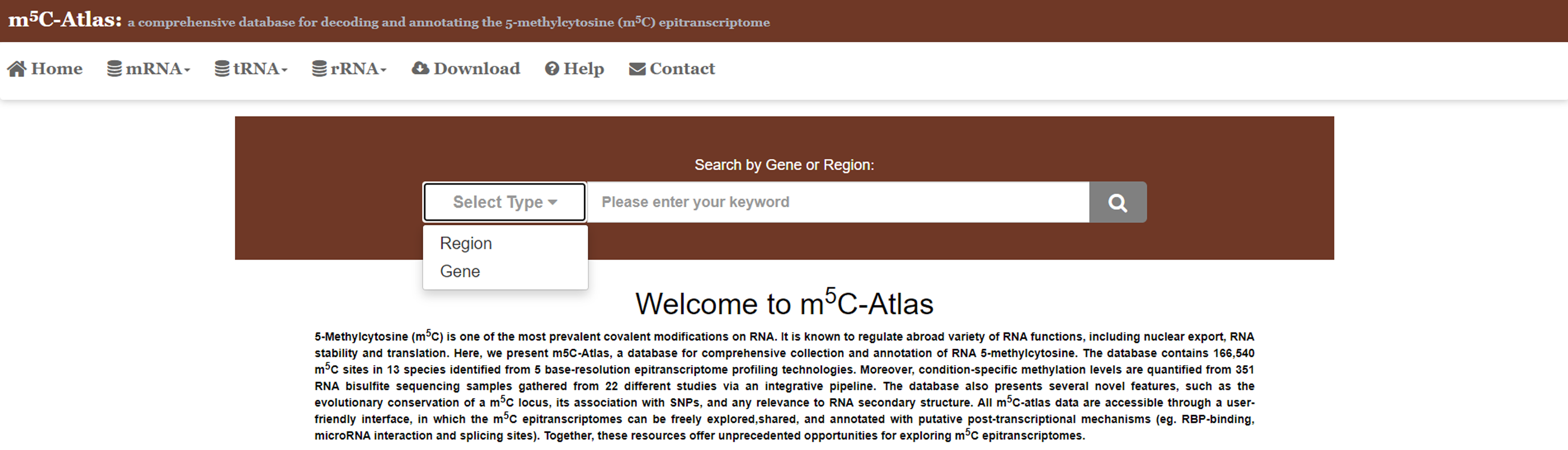
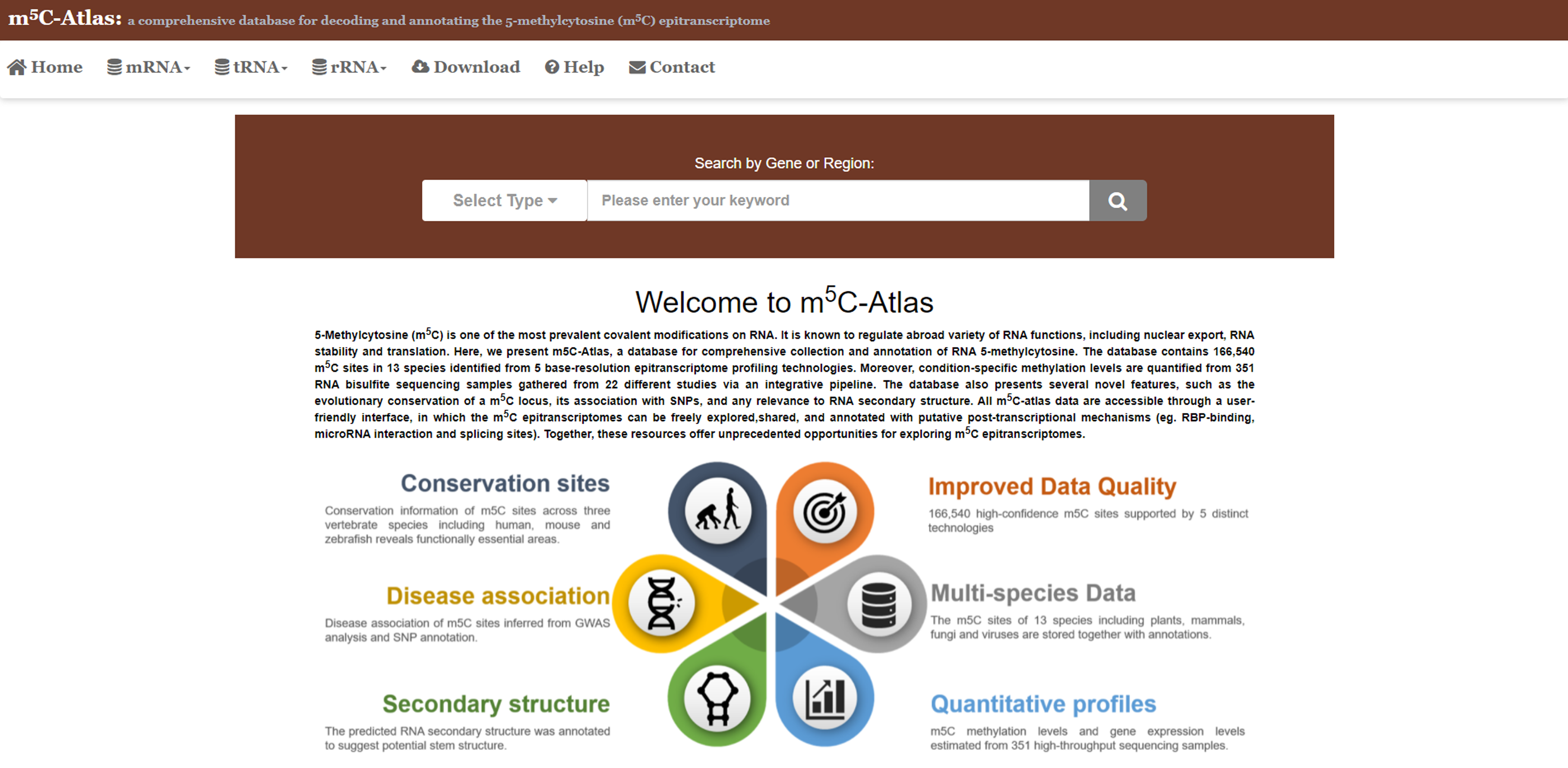 2)Set the query type (i.e., gene name or genomic coordinates) and specify the genes or regions you are interested in. Click the button, then m5C sites locate in the genes or regions will be returned.
2)Set the query type (i.e., gene name or genomic coordinates) and specify the genes or regions you are interested in. Click the button, then m5C sites locate in the genes or regions will be returned.
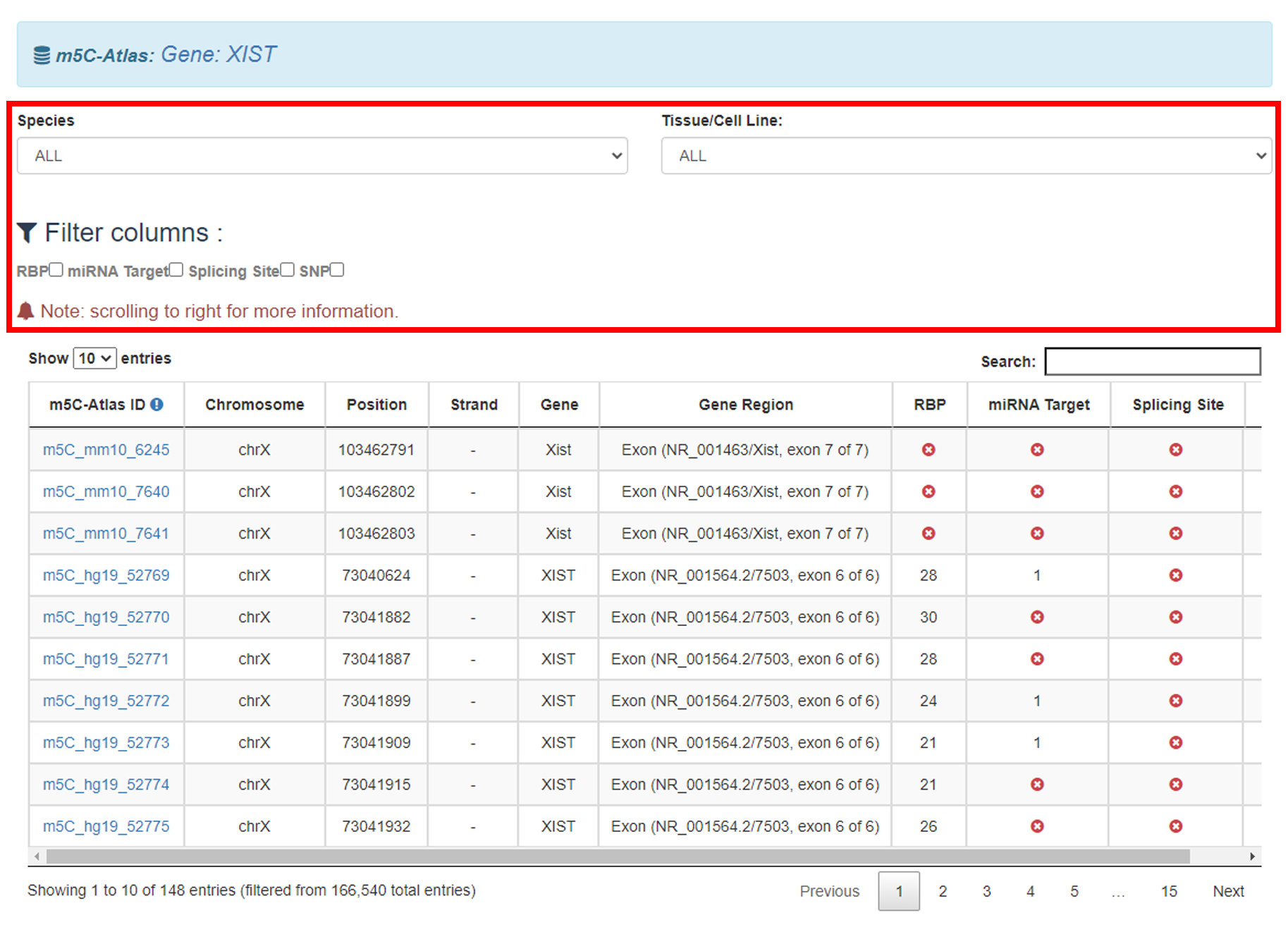
 3)Specify the species, related statistics (i.e., number of m5C, RBP binding sites, miRNA target sites, diseases related SNP sites and conserved sites) and distribution of m5C sites within different tissues will be returned.
3)Specify the species, related statistics (i.e., number of m5C, RBP binding sites, miRNA target sites, diseases related SNP sites and conserved sites) and distribution of m5C sites within different tissues will be returned.
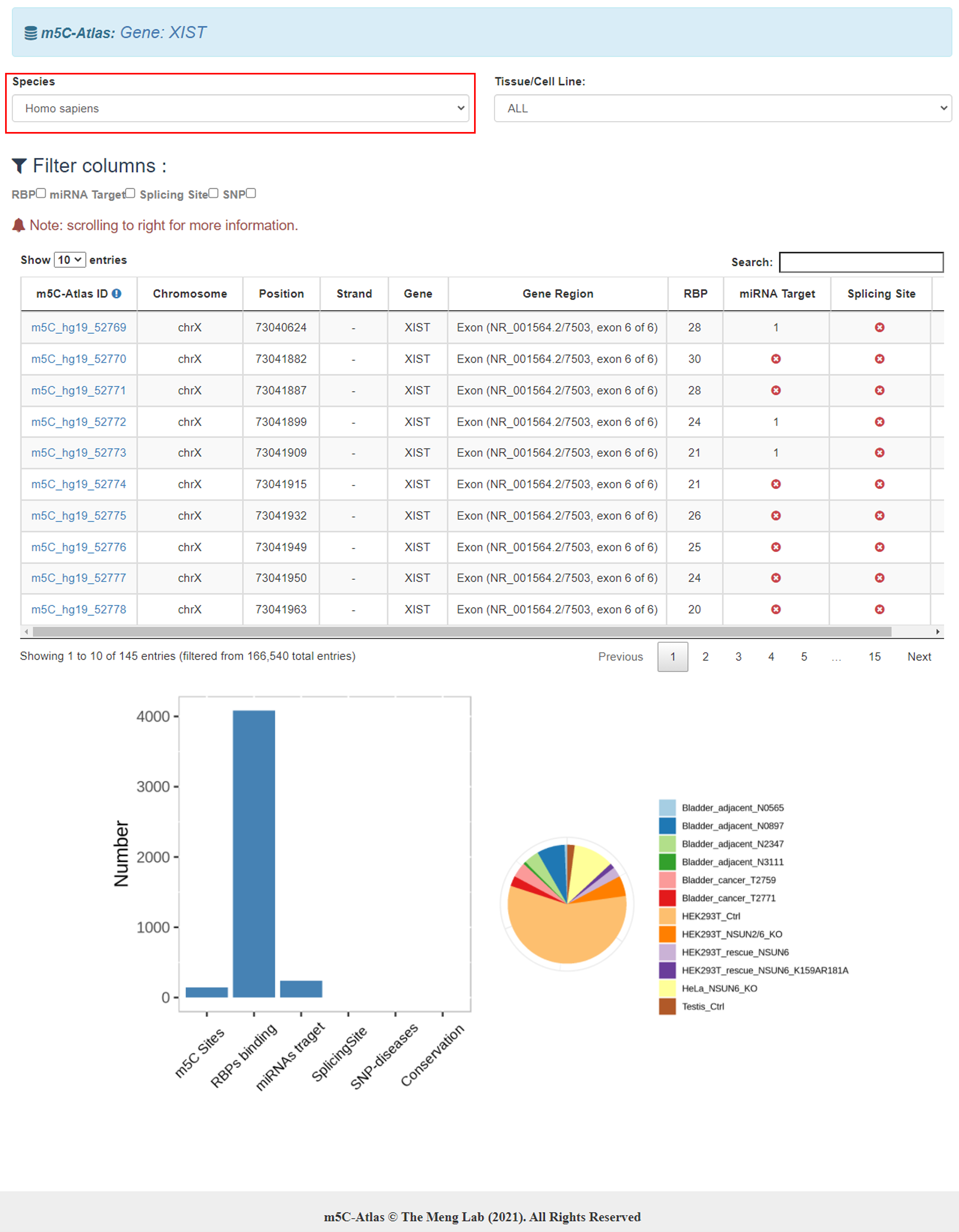 4)Click the m5C-Atlas ID, a new page will be returned, containing the basic site information, conditions of experiments that reported this site, techniques and involved miRNA target, splicing site, disease related SNP and RNA binding proteins (if exist).
4)Click the m5C-Atlas ID, a new page will be returned, containing the basic site information, conditions of experiments that reported this site, techniques and involved miRNA target, splicing site, disease related SNP and RNA binding proteins (if exist).

2.Database
1)m5C-Atlas provides users a browser page to query m5C sites by species and tissues/cell lines.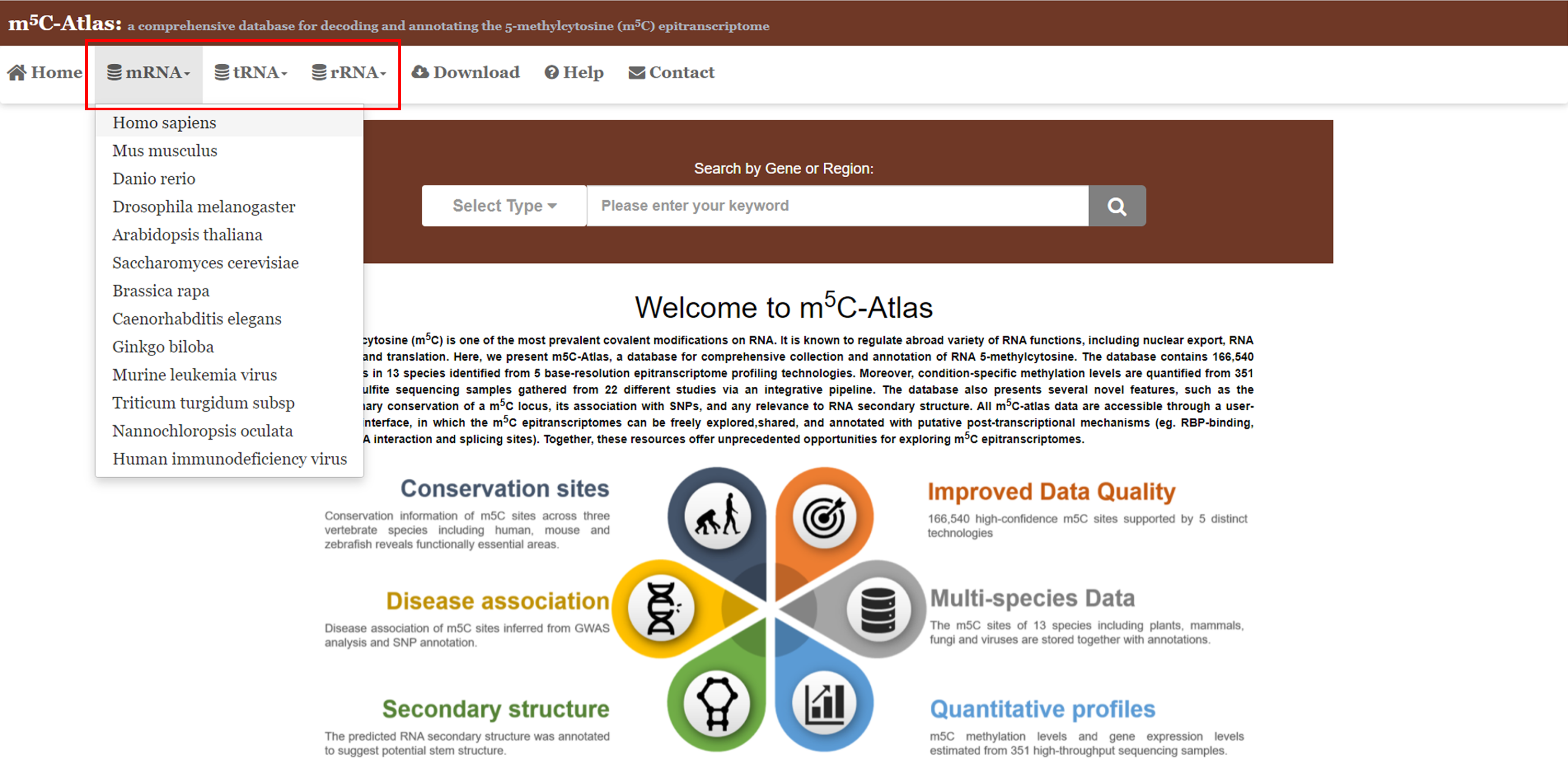 ===============================================
===============================================
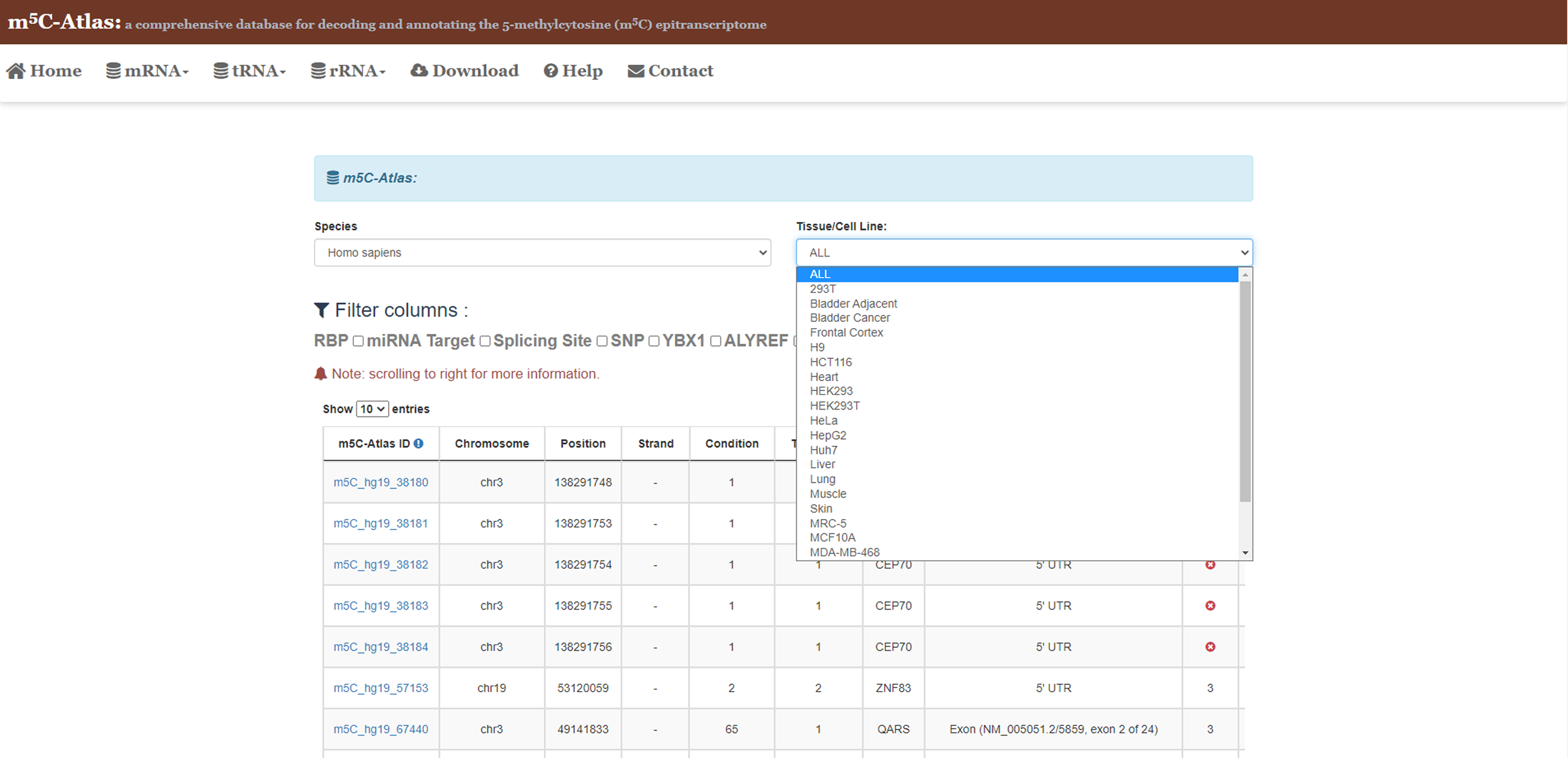 2)Set the species and tissue/cell lines of interest, m5C sites with unique ID will be returned
2)Set the species and tissue/cell lines of interest, m5C sites with unique ID will be returned
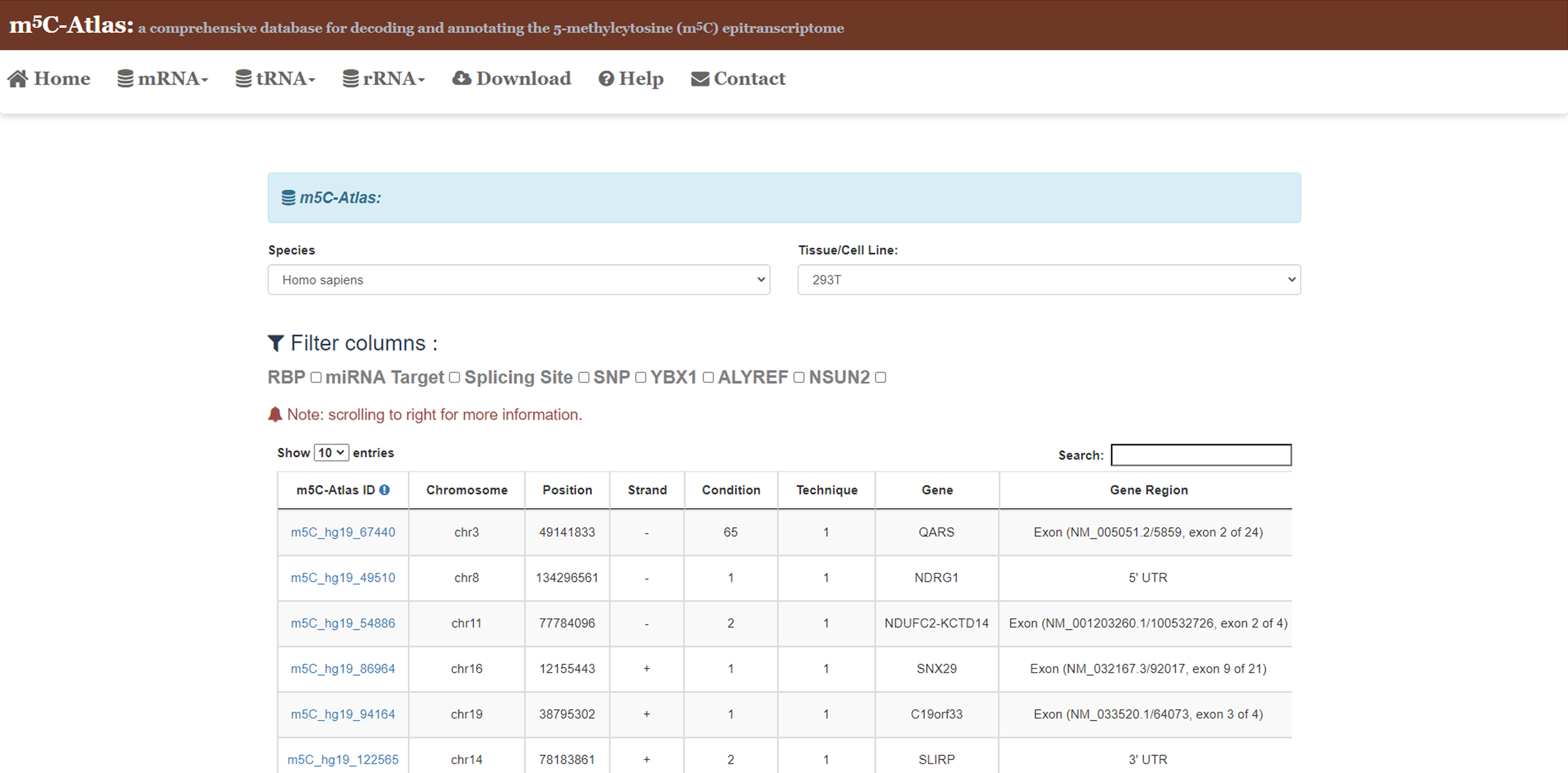 3)Tick the box, results will be filtered by columns.
3)Tick the box, results will be filtered by columns.
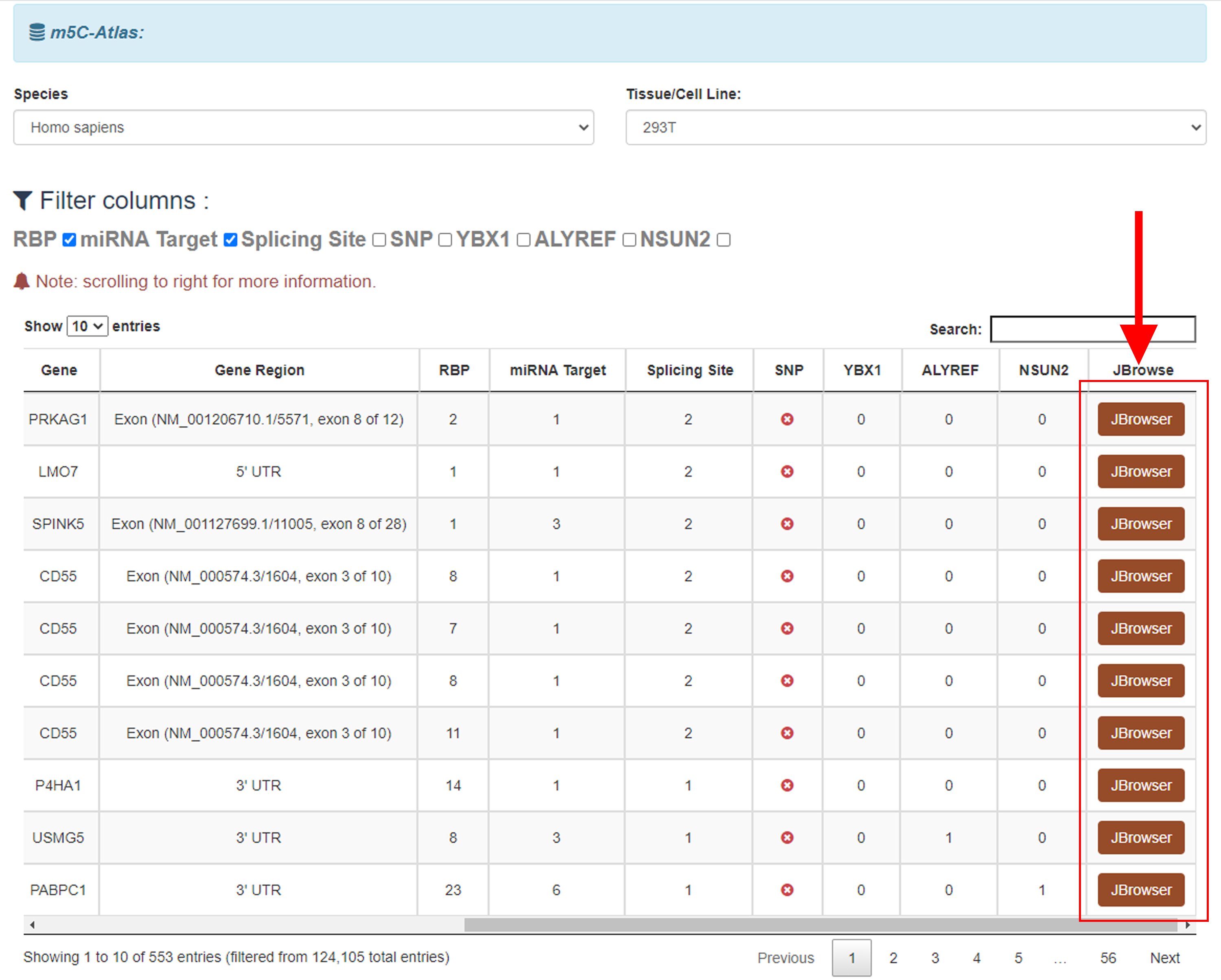
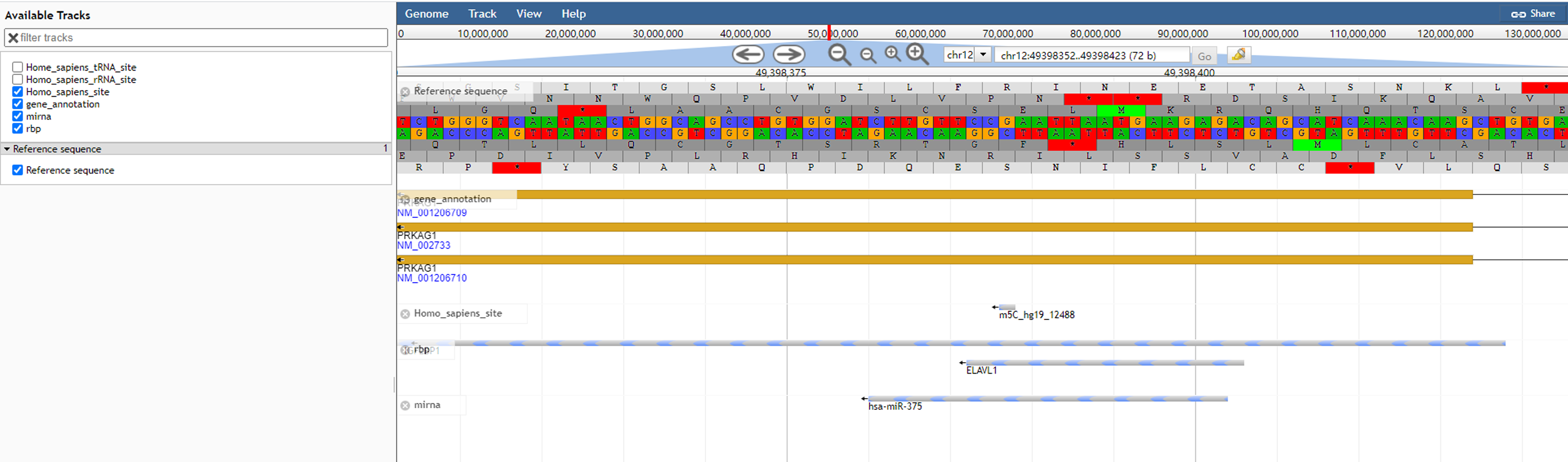
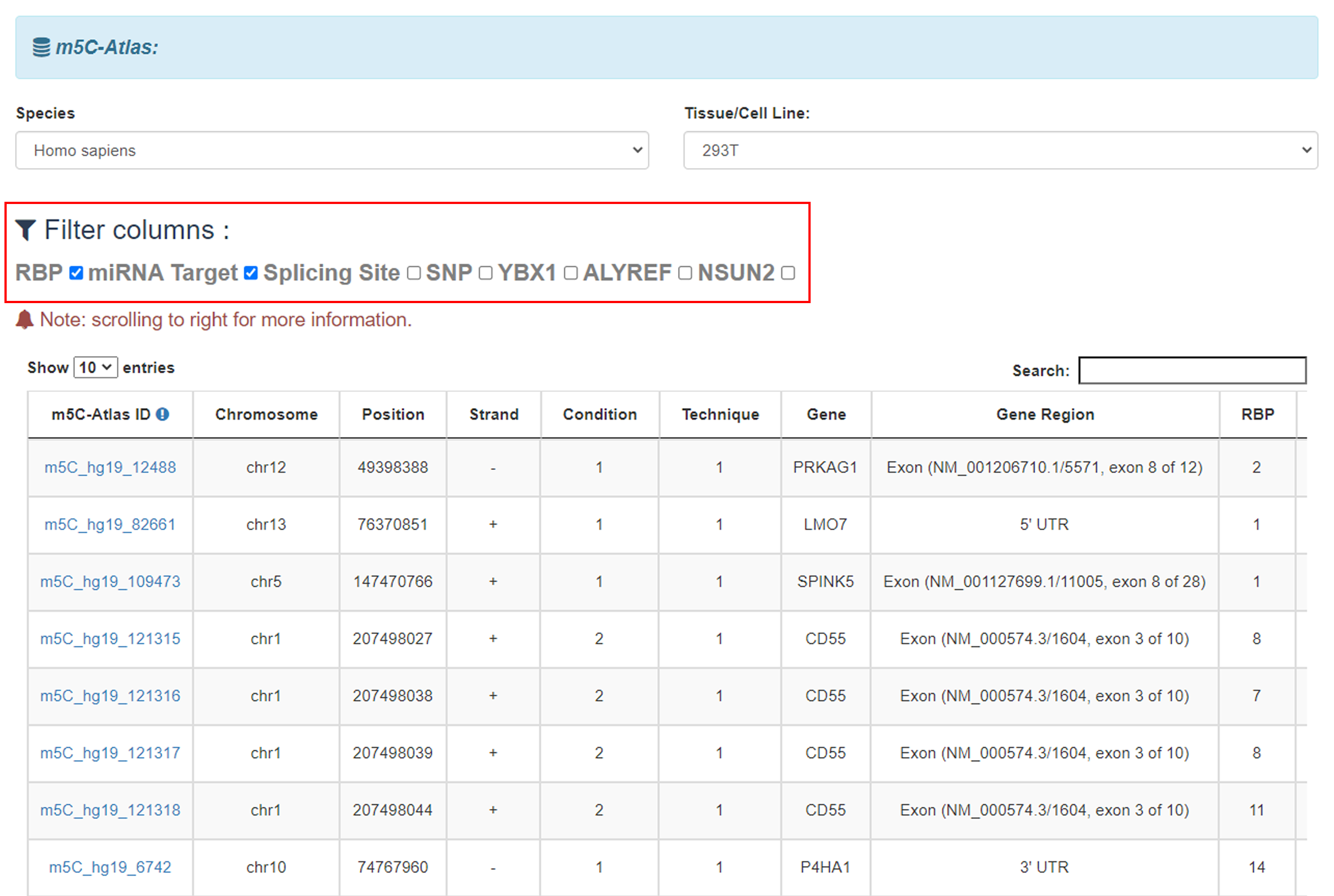 4)Click the Jbrowse button to view modification sites with annotations
4)Click the Jbrowse button to view modification sites with annotations
 ===============================================
===============================================
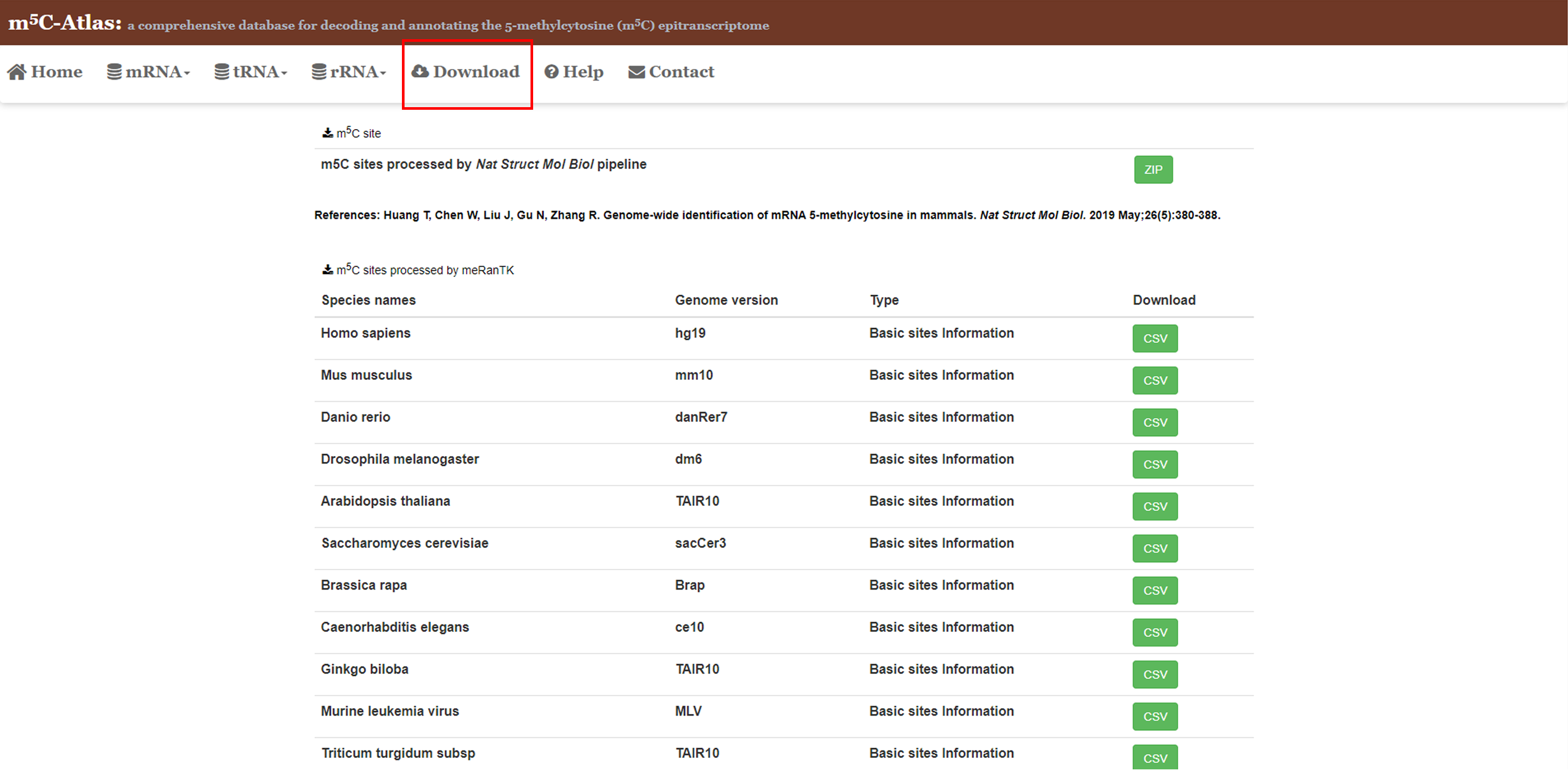
3.Download
1)m5C-Atlas provides users a browser page to download m5C site location information, annotations, technique, conservation and experimental condition information. 2)Click the green button to download CSV files.